21. RNA metabolism
Learning objectives
- What are the differences between RNA and DNA?
- Which shapes can RNA take on?
- Which are the three most important types of RNA, and what are their functions?
- What are the similarities and differences between DNA and RNA polymerase?
- What is the function of the sigma factor of prokaryotic RNA polymerase?
- Describe rho-dependent termination
- Describe rho-independent termination
- What is the mechanism of action of actinomycin D?
- What is the mechanism of action of rifampin?
- What is the mechanism of action of α-amanitin?
- What are the differences between gene transcription in prokaryotes and eukaryotes?
- Describe capping and its function
- Describe polyadenylation and its function
- Describe splicing and the two methods of it
- Describe tRNA processing
RNA
RNA and DNA have some differences, which are described in the table below.
| Property | DNA | RNA |
|---|---|---|
| Bases | Thymine (T), cytosine (C), adenine (A), guanine (G) | Uracil (U), cytosine (C), adenine (A), guanine (G) |
| Modification of bases | Sometimes | Frequently – many modifications are possible |
| Sugar component | Deoxyribose | Ribose |
| Structure | Double-stranded
Double helix |
Single-stranded (with a few exceptions)
Can form hairpins, loops, bulges |
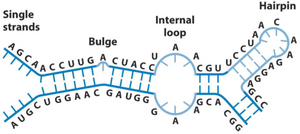
While DNA is always in a double-helix shape, RNA is more flexible regarding shapes. RNA is single-stranded, but can form short double-stranded sections with bulges, internal loops, and hairpin structures, as seen below. These structures give different RNA molecules different functions and properties.
There are many types of RNA molecules, the most important being mRNA, tRNA, and rRNA. Small RNA molecules are described in the next topic.
mRNA (messenger RNA) is the coding RNA. It functions as the middle man between DNA and protein synthesis. After being transcribed by DNA the mRNA will travel to the ribosomes to get translated into protein.
tRNA (transfer RNA) is used in protein synthesis. tRNA transports amino acids to the ribosome, and matches the nucleotide sequence on mRNA to the corresponding amino acid.
rRNA (ribosomal RNA) are components of the ribosomes themselves. They’re ribozymes, meaning that they are RNA molecules with catalytic activity.
RNA synthesis
RNA is synthesised by RNA polymerase, an enzyme similar to DNA polymerase. These enzymes have certain similarities and differences. The similarities are:
- Both need aspartate and Mg2+ to work
- Both synthesize in 5’ -> 3’ direction
- Both have the same mechanism of elongation
- Both have similar processivity
- Both hydrolyse pyrophosphate as part of the elongation mechanism
They have some differences. RNA polymerase does not require a primer and does not have any nuclease activity. The enzyme also always creates a cap on the 5′ end of its product, which we’ll return to later. The most important difference between RNA and DNA polymerase is that RNA polymerase only uses certain genes as its template and not the whole DNA molecule.
When RNA polymerase transcribes DNA, it must first unwind a portion of the coiled DNA into two separate strands. RNA polymerase has intrinsic helicase activity and does not require a separate protein to unwind the DNA.

The DNA is unwinded into to strands, the nontemplate strand (which the new RNA will be equal to) and the template strand, which RNA pol will use as a template. The unwound portion is called the “transcription bubble”. When RNA pol works its way along the DNA molecule, it will unwind new DNA and rewind the DNA it has already transcribed. In this way, the transcription bubble is always moving.
The σ subunit
RNA polymerase has five different subunits in the bacterium E. coli. They are the α, β, β’, σ and ω subunits. The σ subunit recognizes gene promoters and can be switched out with different σ subunits. These subunits are called sigma factors or specificity factors. Each σ subunit recognizes specific promoters for specific genes. Without the σ subunit, RNA polymerase cannot bind a promoter, and therefore cannot transcribe anything.
There exist several σ subunits which can be switched out at any time. The “standard” σ subunit is called σ70, which recognizes and binds “housekeeping” genes, genes that should be expressed when nothing special is happening in the cell.
However, if the cell requires transcription of different genes, a different σ subunit is needed. For example, if the cell is exposed to some sort of stress, for example an increase in temperature, it will switch out the σ70 subunit for a different one, in this case σ32. This subunit recognizes promotors for so-called “heat shock genes”, genes that should be expressed in response to stress. By switching out the σ subunit, the cell can rapidly change what kinds of genes it wants transcribed.
Termination of transcription
We’ve looked at how RNA polymerase begins transcription, but how does it know when to stop transcribing? It can be done in two ways; by the function of a protein called rho-protein (rho-dependent termination) or without it (rho-protein independent termination).
In rho-dependent termination, a protein called rho-protein, which is a helicase, binds to a binding site on the end of the transcribed RNA chain. Rho travels up the RNA chain and unwinds the DNA-RNA hybrid, eventually dislodging RNA polymerase. This requires ATP.
In rho-independent termination, the transcribed RNA molecule creates a hairpin loop on itself, on the end. This hairpin loop forms due to C≡G rich regions in the RNA. This hairpin loop on the end of RNA causes the RNA to dissociate from RNA polymerase. This method does not require ATP.
RNA polymerase inhibitors
Several medications and toxins are RNA polymerase inhibitors. The ones you should know are in the table below.
| Name of compound | Actinomycin D | Rifampicin/rifampin | α-amanitin |
|---|---|---|---|
| Inhibits which RNA polymerase? | All | RNA polymerase | II and III |
| In bacteria or eukaryotes? | Both | Bacteria | Eukaryotes |
| Mechanism of action? | Binds to DNA, blocking RNA polymerase | Binds to beta subunit, blocking initiation of transcription | Blocks mRNA synthesis by binding to RNA pol. |
Transcription in eukaryotes
Transcription in eukaryotes is very similar to in prokaryotes, however, there are differences.
Prokaryotes don’t have nuclei in their cells. This means that DNA and the ribosomes are located at the same place (in the cytosol). Eukaryotes however, have their DNA in the nucleus, separated from the ribosomes. Because transcription must happen where the DNA is, and translation must happen where the ribosomes are, the first difference becomes clear. In prokaryotes, RNA is transcribed and translated at the same time at the same place, while in eukaryotes, RNA is first transcribed in the nucleus, then transported out of the nucleus into the cytoplasm, where it is translated into protein.
Bacteria only have one RNA polymerase, but eukaryotes have three. RNA polymerase I, which is located in the nucleolus and transcribes rRNA, RNA polymerase II, which is located in the nucleus and transcribes mRNA and snRNA, and RNA polymerase III, which is also in the nucleus, but transcribes tRNA and rRNA.
RNA processing
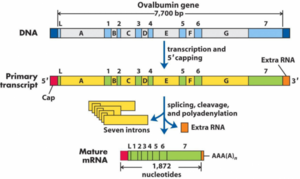
RNA is processed in many ways before it is translated into protein. The most important are capping, tailing and splicing. We call transcribed mRNA immature until it is processed in these manners. At that time, it becomes mature mRNA, ready for translation.
5′ capping
All eukaryotic mRNA has a so-called 5’ cap on the 5′ end. The cap is composed of a 7-methylguanosine and three phosphate groups. The cap protects the mRNA, and facilitates binding to the ribosome.
Tailing/polyadenylation
All eukaryotic mRNA has a so-called poly(A) tail on the 3’-end. This tail is composed of 100-250 adenine residues on the 3’-end of the mRNA. Its function is protection and transport. The tail is created by poly-A polymerase. The reaction is like this (and yes you need to know this):
RNA + nATP —-> RNA-(AMP)n + nPPi
Splicing
Immature mRNA is composed by introns and exons. Introns are sequences that are not meant for translation, so they are “spliced” out (removed). They can be spliced by two mechanisms:
- Splicing by the spliceosome
- Self-splicing
Alternative splicing can yield completely different products, very similar products, or anything in between. This works by splicing mRNAs differently. By varying what we splice out of the immature mRNA and what we leave in, we can change the resulting protein completely. Alternative splicing also increases the number of proteins which can be transcribed. In fact, humans have approximately 33 000 genes, but more than 500 000 different proteins can be transcribed.
The spliceosome is a complex molecule which consists of many small nuclear ribonucleoprotein particles (snRNPs) and the mRNA to be processed. The spliceosome recognizes the borders between the exons and the introns and splices the introns out. The exons are then ligated.
Self-splicing is an alternative method of splicing. Some introns are ribozymes, which catalyse the out-splicing of themselves.
tRNA processing
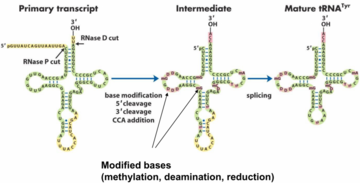
tRNAs are have important functions and are therefore processed in a certain way. There are four important steps in tRNA processing:
- 5′ cleavage
- 3′ cleavage
- Base modification
- CCA addition
After transcription by RNA pol III tRNAs are cut at both the 5’-end and the 3’-end, by RNase P and RNase D respectively. Many of the bases on the tRNA are modified by methylation, deamination and reduction. This yields many unusual bases, like 1-methylguanosine, ribothymidine, pseudouridine, etc.
The nucleotide sequence CCA is added to the 3’-end of the tRNA. This sequence is important, as it’s where the amino acid will bind to.
Summary
- What are the differences between RNA and DNA?
- RNA uses U, DNA uses T
- RNA uses ribose, DNA uses deoxyribose
- RNA is single-stranded, DNA is double-stranded
- Which shapes can RNA take on?
- RNA is single-stranded, but can form short double-stranded sections with bulges, internal loops, and hairpin structures
- Which are the three most important types of RNA, and what are their functions?
- mRNA is the template for protein synthesis
- tRNA transports amino acids to the ribosome, and matches the nucleotide sequence on mRNA to the corresponding amino acid
- rRNA are components of the ribosomes
- What are the similarities and differences between DNA and RNA polymerase?
- Both:
- need aspartate and Mg2+ to work
- synthesize in 5’ -> 3’ direction
- have the same mechanism of elongation
- have similar processivity
- hydrolyse pyrophosphate as part of the elongation mechanism
- Only RNA polymerase:
- does not require a primer
- does not have any nuclease activity
- always creates a cap on the 5′ end of its product
- only uses certain genes as its template and not the whole DNA molecule
- Both:
- What is the function of the sigma factor of prokaryotic RNA polymerase?
- The σ subunit binds to the promoter of the gene.
- By switching out the σ subunit the bacterium can choose which genes to transcribe
- Describe rho-dependent termination
- During transcription a protein called Rho binds to the “tail” of the RNA polymerase when it begins transcription. Rho travels up this tail towards the “body” of RNA pol, and when it reaches it, it will separate the RNA from the RNA polymerase enzyme, terminating the transcription.
- Describe rho-independent termination
- The transcribed RNA molecule creates a hairpin loop on itself, on the end. This hairpin loop on the end of RNA causes the RNA to dissociate from RNA polymerase.
- What is the mechanism of action of actinomycin D?
- Actinomycin D binds to both prokaryotic and eukaryotic DNA, blocking RNA polymerase from transcribing
- What is the mechanism of action of rifampin?
- Rifampin binds to the beta subunit of prokaryotic RNA polymerase, preventing initiation of transcription
- What is the mechanism of action of α-amanitin?
- α-amanitin binds to eukaryotic RNA polymerase II and III and inhibits them
- What are the differences between gene transcription in prokaryotes and eukaryotes?
- In prokaryotes transcription and translation occurs simultaneously and at the same site
- In eukaryotes transcription and translation occurs at different times at different sites
- Bacteria only have one RNA polymerase, but eukaryotes have three. RNA polymerase I, which is located in the nucleolus and transcribes rRNA, RNA polymerase II, which is located in the nucleus and transcribes mRNA and snRNA, and RNA polymerase III, which is also in the nucleus, but transcribes tRNA and rRNA.
- Describe capping and its function
- Capping refers to RNA polymerase putting a 7-methylguanosine cap on the 5′ end of RNA
- The cap protects the mRNA, and facilitates binding to the ribosome
- Describe polyadenylation and its function
- Polyadenylation refers to putting many adenine residues on the 3′ end of the RNA
- This is catalysed by poly-A polymerase
- The tail’s function is protection and transport
- Describe splicing and the two methods of it
- Splicing refers to removing introns from immature mRNA, leaving only exons
- The spliceosome is a molecular complex which consists of many small nuclear ribonucleoprotein particles (snRNPs) and the mRNA to be processed. The spliceosome recognizes the borders between the exons and the introns and splices the introns out. The exons are then ligated.
- Self-splicing is an alternative method of splicing. Some introns are ribozymes, which catalyse the out-splicing of themselves.
- Describe tRNA processing
- 5′ cleavage by RNase P
- 3′ cleavage by RNAse D
- Base modification by methylation, deamination and reduction
- CCA addition
- The CCA region will bind to the amino acid
